Lecture 8: Xarray for multidimensional gridded data¶
In last week's lecture, we saw how Pandas provided a way to keep track of additional "metadata" surrounding tabular datasets, including "indexes" for each row and labels for each column. These features, together with Pandas' many useful routines for all kinds of data munging and analysis, have made Pandas one of the most popular python packages in the world.
However, not all earth and environmental science datasets easily fit into the "tabular" model (i.e. rows and columns) imposed by Pandas. In particular, we often deal with multidimensional data. By multidimensional data (also often called N-dimensional), I mean data with many independent dimensions or axes. For example, we might represent Earth's surface temperature $T$ as a three dimensional variable
$$ T(x, y, t) $$
where $x$ is longitude, $y$ is latitude, and $t$ is time.
The point of xarray is to provide pandas-level convenience for working with this type of data.
Xarray Data Model¶
Here is an example of how we might structure a dataset for a weather forecast:
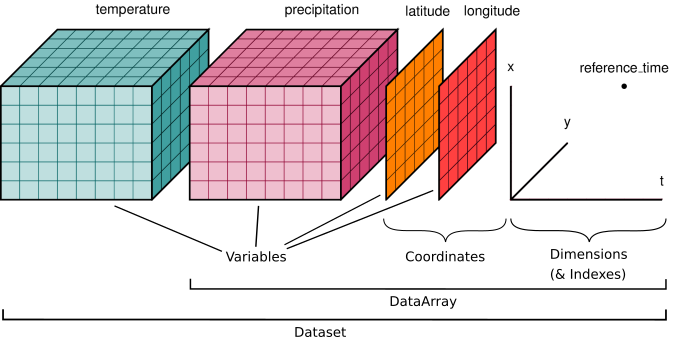
You'll notice multiple data variables (temperature, precipitation), coordinate variables (latitude, longitude), and dimensions (x, y, t). We'll cover how these fit into Xarray's data structures below.
Xarray functionality¶
Xarray doesn’t just keep track of labels on arrays – it uses them to provide a powerful and concise interface. For example:
Apply operations over dimensions by name:
x.sum('time').Select values by label (or logical location) instead of integer location:
x.loc['2014-01-01']orx.sel(time='2014-01-01').Mathematical operations (e.g.,
x - y) vectorize across multiple dimensions (array broadcasting) based on dimension names, not shape.Easily use the split-apply-combine paradigm with groupby:
x.groupby('time.dayofyear').mean().Database-like alignment based on coordinate labels that smoothly handles missing values:
x, y = xr.align(x, y, join='outer').Keep track of arbitrary metadata in the form of a Python dictionary:
x.attrs.
The N-dimensional nature of xarray’s data structures makes it suitable for
dealing with multi-dimensional scientific data, and its use of dimension names
instead of axis labels (dim='time' instead of axis=0) makes such arrays much
more manageable than the raw numpy ndarray: with xarray, you don’t need to keep
track of the order of an array’s dimensions or insert dummy dimensions of size 1
to align arrays (e.g., using np.newaxis).
The immediate payoff of using xarray is that you’ll write less code. The long-term payoff is that you’ll understand what you were thinking when you come back to look at it weeks or months later.
Xarray data structures¶
Like Pandas, xarray has two fundamental data structures:
- a
DataArray, which holds a single multi-dimensional variable and its coordinates - a
Dataset, which holds multiple variables that potentially share the same coordinates
DataArray¶
A DataArray has four essential attributes:
values: anumpy.ndarrayholding the array’s valuesdims: dimension names for each axis (e.g.,('x', 'y', 'z'))coords: a dict-like container of arrays (coordinates) that label each point (e.g., 1-dimensional arrays of numbers, datetime objects or strings)attrs: anOrderedDictto hold arbitrary metadata (attributes)
Let's start by constructing some DataArrays manually
Install Xarray:¶
$ conda install -c conda-forge xarray dask netCDF4 bottleneck python-graphviz
# First import xarray
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
Xarray Data Structure¶
Xarray has a few small real-world tutorial datasets hosted in this GitHub repository https://github.com/pydata/xarray-data.
We'll use the xarray.tutorial.load_dataset convenience function to download and open the air_temperature (National Centers for Environmental Prediction) Dataset by name.
ds = xr.tutorial.load_dataset("air_temperature")
ds
<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 296.5 296.2 295.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...We can access "layers" of the Dataset (individual DataArrays) with dictionary syntax
ds["air"]
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]We can save some typing by using the "attribute" or "dot" notation. This won't work for variable names that clash with built-in method names (for example, mean).
ds.air
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]What is included in xarray Dataset?¶
The output consists of:
- a summary of all dimensions of the
Dataset(lat: 25, time: 2920, lon: 53): this tells us that the first dimension is namedlatand has a size of25, the second dimension is namedtimeand has a size of2920, and the third dimension is namedlonand has a size of53. Because we will access the dimensions by name, the order doesn't matter. - an unordered list of coordinates or dimensions with coordinates with one item per line. Each item has a name, one or more dimensions in parentheses, a dtype and a preview of the values.
- an alphabetically sorted list of dimensions without coordinates (if there are any)
- an unordered list of attributes, or metadata
DataArray¶
The DataArray class consists of an array (data) and its associated dimension names, labels, and attributes (metadata).
da = ds["air"]
da
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]We can also access the data array directly:
ds.air.data
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...,
[296.4 , 295.9 , 296.19998, ..., 295.4 , 295.1 ,
294.79 ],
[296.19998, 296.69998, 296.79 , ..., 295.6 , 295.5 ,
295.1 ],
[296.29 , 297.19998, 297.4 , ..., 296.4 , 296.4 ,
296.6 ]],
[[242.29999, 242.2 , 242.29999, ..., 234.29999, 236.09999,
238.7 ],
[244.59999, 244.39 , 244. , ..., 230.29999, 232. ,
235.7 ],
[256.19998, 255.5 , 254.2 , ..., 231.2 , 233.2 ,
238.2 ],
...,
[295.6 , 295.4 , 295.4 , ..., 296.29 , 295.29 ,
295. ],
[296.19998, 296.5 , 296.29 , ..., 296.4 , 296. ,
295.6 ],
[296.4 , 296.29 , 296.4 , ..., 297. , 297. ,
296.79 ]],
...,
[[243.48999, 242.98999, 242.09 , ..., 244.18999, 244.48999,
244.89 ],
[249.09 , 248.98999, 248.59 , ..., 240.59 , 241.29 ,
242.68999],
[262.69 , 262.19 , 261.69 , ..., 239.39 , 241.68999,
245.18999],
...,
[294.79 , 295.29 , 297.49 , ..., 295.49 , 295.38998,
294.69 ],
[296.79 , 297.88998, 298.29 , ..., 295.49 , 295.49 ,
294.79 ],
[298.19 , 299.19 , 298.79 , ..., 296.09 , 295.79 ,
295.79 ]],
[[245.79 , 244.79 , 243.48999, ..., 243.29 , 243.98999,
244.79 ],
[249.89 , 249.29 , 248.48999, ..., 241.29 , 242.48999,
244.29 ],
[262.38998, 261.79 , 261.29 , ..., 240.48999, 243.09 ,
246.89 ],
...,
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Named dimensions¶
.dims are the named axes of your data. They may (dimension coordinates) or may not (dimensions without coordinates) have associated values.
In this case we have 2 spatial dimensions (latitude and longitude are stored with shorthand names lat and lon) and one temporal dimension (time).
ds.air.dims
('time', 'lat', 'lon')
Coordinates¶
.coords is a simple dict-like data container
for mapping coordinate names to values.
Here we see the actual timestamps and spatial positions of our air temperature data:
ds.air.coords
Coordinates: * lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0 * lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0 * time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes¶
.attrs is a dictionary that can contain arbitrary Python objects (strings, lists, integers, dictionaries, etc.) containing information about your data. Your only
limitation is that some attributes may not be writeable to certain file formats.
ds.air.attrs
{'long_name': '4xDaily Air temperature at sigma level 995',
'units': 'degK',
'precision': 2,
'GRIB_id': 11,
'GRIB_name': 'TMP',
'var_desc': 'Air temperature',
'dataset': 'NMC Reanalysis',
'level_desc': 'Surface',
'statistic': 'Individual Obs',
'parent_stat': 'Other',
'actual_range': array([185.16, 322.1 ], dtype='>f4')}
# assign your own attributes!
ds.air.attrs["new_attr"] = "xarray"
ds.air.attrs
{'long_name': '4xDaily Air temperature at sigma level 995',
'units': 'degK',
'precision': 2,
'GRIB_id': 11,
'GRIB_name': 'TMP',
'var_desc': 'Air temperature',
'dataset': 'NMC Reanalysis',
'level_desc': 'Surface',
'statistic': 'Individual Obs',
'parent_stat': 'Other',
'actual_range': array([185.16, 322.1 ], dtype='>f4'),
'new_attr': 'xarray'}
Working with Labelled Data¶
Xarray's labels make working with multidimensional data much easier. Metadata provides context and provides code that is more legible. This reduces the likelihood of errors from typos and makes analysis more intuitive and fun!
## Without Xarray, let's try to use matplotlib to plot the data
temp = ds.air.data
plt.pcolormesh(temp[0,:,:])
<matplotlib.collections.QuadMesh at 0x1945afee0>
### Add Lat and Lon
temp = ds.air.data
lat = ds.air.lat.data
lon = ds.air.lon.data
plt.pcolormesh(lon, lat, temp[0,:,:])
<matplotlib.collections.QuadMesh at 0x1947868b0>
temp.mean(axis=1) ## what did I just do? I can't tell by looking at this line.
array([[279.39798, 279.6664 , 279.66122, ..., 279.9508 , 280.31522,
280.6624 ],
[279.05722, 279.538 , 279.7296 , ..., 279.77563, 280.27002,
280.79764],
[279.0104 , 279.2808 , 279.5508 , ..., 279.682 , 280.19763,
280.81403],
...,
[279.63 , 279.934 , 280.534 , ..., 279.802 , 280.346 ,
280.77798],
[279.398 , 279.66602, 280.31796, ..., 279.766 , 280.34198,
280.834 ],
[279.27 , 279.354 , 279.88202, ..., 279.42596, 279.96997,
280.48196]], dtype=float32)
ds.air.isel(time=0).plot()
<matplotlib.collections.QuadMesh at 0x194849940>
ds.air.mean(dim="time").plot()
<matplotlib.collections.QuadMesh at 0x194942490>
Selecting Data (Indexing)¶
We can always use regular numpy indexing and slicing on DataArrays
Position-based Indexing¶
Indexing a DataArray directly works (mostly) just like it does for numpy ndarrays, except that the returned object is always another DataArray:
This approach however does not take advantage of the dimension names and coordinate location information that is present in a Xarray object.
da[:, 20, 40]
<xarray.DataArray 'air' (time: 2920)>
array([295. , 294.4 , 294.5 , ..., 297.29, 297.79, 297.99], dtype=float32)
Coordinates:
lat float32 25.0
lon float32 300.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarrayda[0,0,0]
<xarray.DataArray 'air' ()>
array(241.2, dtype=float32)
Coordinates:
lat float32 75.0
lon float32 200.0
time datetime64[ns] 2013-01-01
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarrayPositional Indexing Using Dimension Names¶
Remembering the axis order can be challenging even with 2D arrays:
- is
np_array[0,3]the first row and third column or first column and third row? - or did I store these samples by row or by column when I saved the data?!.
The difficulty is compounded with added dimensions.
Xarray objects eliminate much of the mental overhead by allowing indexing using dimension names instead of axes numbers:
da.isel(lat=0, lon=0).plot();
Slicing is also possible similarly:
da.isel(time=slice(0, 20), lat=0, lon=0).plot();
{note}
Using the `isel` method, the user can choose/slice the specific elements from a Dataset or DataArray.
So far, we have explored positional indexing, which relies on knowing the exact indices. But, what if you wanted to select data specifically for a particular latitude? It becomes challenging to determine the corresponding indices in such cases. Xarray reduce this complexity by introducing label-based indexing.
Label-based Indexing¶
To select data by coordinate labels instead of integer indices we can use sel instead of isel:
For example, let's select all data for Lat 25 °N and Lon 210 °E using sel :
da.sel(lat=25, lon=210).plot();
Similarly we can do slicing or filter a range using the .slice function:
# demonstrate slicing
da.sel(lon=slice(210, 215))
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 3)>
array([[[244.09999, 243.89 , 243.59999],
[243.39 , 242.39 , 241.7 ],
[246. , 244.39 , 243.09999],
...,
[295.5 , 294. , 293.6 ],
[296.1 , 295.1 , 294.6 ],
[296.9 , 296.4 , 296. ]],
[[243.59999, 243.79999, 244. ],
[243.7 , 243.29999, 242.79999],
[249.29999, 247.5 , 245.5 ],
...,
[295.1 , 294.1 , 293.79 ],
[296.1 , 295.6 , 295.1 ],
[297.1 , 296.69998, 296.4 ]],
[[242.89 , 243.59999, 244.5 ],
[242.79999, 242.39 , 242.29999],
[250.2 , 248.09999, 246.29999],
...,
...
...,
[297.99 , 297.49 , 297.29 ],
[298.59 , 298.49 , 298.19 ],
[299.29 , 298.99 , 298.59 ]],
[[240.29 , 238.89 , 237.79 ],
[245.68999, 243.98999, 242.29 ],
[259.38998, 257.69 , 255.48999],
...,
[297.79 , 297.49 , 297.49 ],
[298.29 , 298.09 , 297.79 ],
[298.79 , 298.69 , 298.49 ]],
[[241.09 , 240.09 , 239.39 ],
[245.09 , 243.09 , 241.09 ],
[257.79 , 254.98999, 251.98999],
...,
[297.29 , 297.29 , 297.38998],
[298.19 , 298.09 , 297.79 ],
[298.88998, 298.69 , 298.38998]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 210.0 212.5 215.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarray# demonstrate slicing
da.sel(lat=slice(25, 50))
<xarray.DataArray 'air' (time: 2920, lat: 0, lon: 53)>
array([], shape=(2920, 0, 53), dtype=float32)
Coordinates:
* lat (lat) float32
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarray# demonstrate slicing
da.sel(lat=slice(50, 25), lon=slice(210, 215))
<xarray.DataArray 'air' (time: 2920, lat: 11, lon: 3)>
array([[[279.5 , 280.1 , 280.6 ],
[279.4 , 280.29 , 281.29 ],
[280.29 , 282. , 283.29 ],
...,
[294.5 , 294.9 , 293.5 ],
[295.4 , 294.69998, 293.19998],
[295.4 , 294. , 292.9 ]],
[[278.4 , 279.19998, 280.6 ],
[279.1 , 279.69998, 280.79 ],
[280.1 , 280.6 , 281.69998],
...,
[294. , 294.5 , 294.19998],
[295.29 , 294.79 , 293.6 ],
[295.6 , 294.29 , 292.9 ]],
[[277.5 , 277.6 , 278.79 ],
[278.79 , 278.29 , 279.1 ],
[280.5 , 279.6 , 279.69998],
...,
...
...,
[292.99 , 292.79 , 293.19 ],
[294.29 , 294.09 , 294.29 ],
[295.49 , 295.19 , 294.69 ]],
[[279.38998, 280.09 , 281.99 ],
[280.49 , 282.59 , 284.29 ],
[283.38998, 285.38998, 285.99 ],
...,
[292.59 , 292.88998, 293.29 ],
[293.88998, 293.88998, 293.99 ],
[295.19 , 295.29 , 294.49 ]],
[[279.88998, 281.79 , 283.69 ],
[281.69 , 283.09 , 283.79 ],
[283.59 , 284.79 , 284.69 ],
...,
[293.59 , 293.59 , 293.49 ],
[294.99 , 294.59 , 294.19 ],
[295.49 , 295.59 , 295.09 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 50.0 47.5 45.0 42.5 40.0 ... 35.0 32.5 30.0 27.5 25.0
* lon (lon) float32 210.0 212.5 215.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarrayDropping using drop_sel¶
If instead of selecting data we want to drop it, we can use drop_sel method with syntax similar to sel:
da
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)>
array([[[241.2 , 242.5 , 243.5 , ..., 232.79999, 235.5 ,
238.59999],
[243.79999, 244.5 , 244.7 , ..., 232.79999, 235.29999,
239.29999],
[250. , 249.79999, 248.89 , ..., 233.2 , 236.39 ,
241.7 ],
...,
[296.6 , 296.19998, 296.4 , ..., 295.4 , 295.1 ,
294.69998],
[295.9 , 296.19998, 296.79 , ..., 295.9 , 295.9 ,
295.19998],
[296.29 , 296.79 , 297.1 , ..., 296.9 , 296.79 ,
296.6 ]],
[[242.09999, 242.7 , 243.09999, ..., 232. , 233.59999,
235.79999],
[243.59999, 244.09999, 244.2 , ..., 231. , 232.5 ,
235.7 ],
[253.2 , 252.89 , 252.09999, ..., 230.79999, 233.39 ,
238.5 ],
...
[293.69 , 293.88998, 295.38998, ..., 295.09 , 294.69 ,
294.29 ],
[296.29 , 297.19 , 297.59 , ..., 295.29 , 295.09 ,
294.38998],
[297.79 , 298.38998, 298.49 , ..., 295.69 , 295.49 ,
295.19 ]],
[[245.09 , 244.29 , 243.29 , ..., 241.68999, 241.48999,
241.79 ],
[249.89 , 249.29 , 248.39 , ..., 239.59 , 240.29 ,
241.68999],
[262.99 , 262.19 , 261.38998, ..., 239.89 , 242.59 ,
246.29 ],
...,
[293.79 , 293.69 , 295.09 , ..., 295.29 , 295.09 ,
294.69 ],
[296.09 , 296.88998, 297.19 , ..., 295.69 , 295.69 ,
295.19 ],
[297.69 , 298.09 , 298.09 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarrayda.drop_sel(lat=75.0, lon=200.0)
<xarray.DataArray 'air' (time: 2920, lat: 24, lon: 52)>
array([[[244.5 , 244.7 , 244.2 , ..., 232.79999, 235.29999,
239.29999],
[249.79999, 248.89 , 247.5 , ..., 233.2 , 236.39 ,
241.7 ],
[267.1 , 267.1 , 266.69998, ..., 249.2 , 253.09999,
256.9 ],
...,
[296.19998, 296.4 , 296.5 , ..., 295.4 , 295.1 ,
294.69998],
[296.19998, 296.79 , 296.5 , ..., 295.9 , 295.9 ,
295.19998],
[296.79 , 297.1 , 297. , ..., 296.9 , 296.79 ,
296.6 ]],
[[244.09999, 244.2 , 244.09999, ..., 231. , 232.5 ,
235.7 ],
[252.89 , 252.09999, 250.79999, ..., 230.79999, 233.39 ,
238.5 ],
[269.4 , 268.6 , 267.4 , ..., 247.2 , 250.5 ,
254.39 ],
...
[293.88998, 295.38998, 297.19 , ..., 295.09 , 294.69 ,
294.29 ],
[297.19 , 297.59 , 297.88998, ..., 295.29 , 295.09 ,
294.38998],
[298.38998, 298.49 , 298.59 , ..., 295.69 , 295.49 ,
295.19 ]],
[[249.29 , 248.39 , 246.98999, ..., 239.59 , 240.29 ,
241.68999],
[262.19 , 261.38998, 259.99 , ..., 239.89 , 242.59 ,
246.29 ],
[272.09 , 271.99 , 271.59 , ..., 255.39 , 258.99 ,
262.49 ],
...,
[293.69 , 295.09 , 296.69 , ..., 295.29 , 295.09 ,
294.69 ],
[296.88998, 297.19 , 297.49 , ..., 295.69 , 295.69 ,
295.19 ],
[298.09 , 298.09 , 298.49 , ..., 296.49 , 296.19 ,
295.69 ]]], dtype=float32)
Coordinates:
* lat (lat) float32 72.5 70.0 67.5 65.0 62.5 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 202.5 205.0 207.5 210.0 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarraySo far, all the above will require us to specify exact coordinate values, but what if we don't have the exact values? We can use nearest neighbor lookups to address this issue:
Nearest Neighbor Lookups¶
The label based selection methods sel() support method and tolerance keyword argument. The method parameter allows for enabling nearest neighbor (inexact) lookups by use of the methods ffill (propagate last valid index forward), backfill or nearest:
da.sel(lat=52.25, lon=251.8998, method="nearest")
<xarray.DataArray 'air' (time: 2920)>
array([262.69998, 263.19998, 270.9 , ..., 264.19 , 265.19 ,
266.99 ], dtype=float32)
Coordinates:
lat float32 52.5
lon float32 252.5
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarrayda.sel(lat=52.25, lon=251.8998, method="ffill")
<xarray.DataArray 'air' (time: 2920)>
array([269.5 , 269.29 , 273.69998, ..., 267.49 , 269.29 ,
268.69 ], dtype=float32)
Coordinates:
lat float32 52.5
lon float32 250.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarraytolerance argument limits the maximum distance for valid matches with an inexact lookup:
da.sel(lat=52.25, lon=251.8998, method="nearest", tolerance=2)
<xarray.DataArray 'air' (time: 2920)>
array([262.69998, 263.19998, 270.9 , ..., 264.19 , 265.19 ,
266.99 ], dtype=float32)
Coordinates:
lat float32 52.5
lon float32 252.5
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarrayda.sel(lat=52.25, lon=198, method="nearest", tolerance=2)
<xarray.DataArray 'air' (time: 2920)>
array([276.69998, 275.79 , 275.29 , ..., 277.49 , 276.79 ,
276.88998], dtype=float32)
Coordinates:
lat float32 52.5
lon float32 200.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]
new_attr: xarray{tip}
All of these indexing methods work on the dataset too!
We can also use these methods to index all variables in a dataset simultaneously, returning a new dataset:
ds.sel(lat=52.25, lon=251.8998, method="nearest")
<xarray.Dataset>
Dimensions: (time: 2920)
Coordinates:
lat float32 52.5
lon float32 252.5
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time) float32 262.7 263.2 270.9 274.1 ... 261.6 264.2 265.2 267.0
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...Datetime Indexing¶
Datetime indexing is a critical feature when working with time series data, which is a common occurrence in atmospheric and environmental sciences. Essentially, datetime indexing allows you to select data points or a series of data points that correspond to certain date or time criteria. This becomes essential for time-series analysis where the date or time information associated with each data point can be as critical as the data point itself.
Let's see some of the techniques to perform datetime indexing in Xarray:
Selecting data based on single datetime¶
Let's say we have a Dataset ds and we want to select data at a particular date and time, for instance, '2013-01-01' at 6AM. We can do this by using the sel (select) method, like so:
ds.sel(time='2013-01-01 06:00')
<xarray.Dataset>
Dimensions: (lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
time datetime64[ns] 2013-01-01T06:00:00
Data variables:
air (lat, lon) float32 242.1 242.7 243.1 243.4 ... 296.4 296.4 296.6
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...By default, datetime selection will return a range of values that match the provided string. For e.g. time="2013-01-01" will return all timestamps for that day (4 of them here):
ds.sel(time='2013-01-01')
<xarray.Dataset>
Dimensions: (lat: 25, time: 4, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2013-01-01T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 297.8 298.0 297.9
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...We can use this feature to select all points in a year:
ds.sel(time="2014")
<xarray.Dataset>
Dimensions: (lat: 25, time: 1460, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2014-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 252.3 251.2 250.0 ... 296.5 296.2 295.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...or a month:
ds.sel(time="2014-May")
# ds.sel(time="2014-05")
<xarray.Dataset>
Dimensions: (lat: 25, time: 124, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2014-05-01 ... 2014-05-31T18:00:00
Data variables:
air (time, lat, lon) float32 264.9 265.0 265.0 ... 296.5 296.2 296.2
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...Selecting data for a range of dates¶
Now, let's say we want to select data between a certain range of dates. We can still use the sel method, but this time we will combine it with slice:
# This will return a subset of the dataset corresponding to the entire year of 2013.
ds.sel(time=slice('2013-01-01', '2013-12-31'))
<xarray.Dataset>
Dimensions: (lat: 25, time: 1460, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2013-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 296.1 295.1 294.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...The slice function takes two arguments, start and stop, to make a slice that includes these endpoints. When we use slice with the sel method, it provides an efficient way to select a range of dates. The above example shows the usage of slice for datetime indexing.
Indexing with a DatetimeIndex or date string list¶
Another technique is to use a list of datetime objects or date strings for indexing. For example, you could select data for specific, non-contiguous dates like this:
dates = ['2013-07-09', '2013-10-11', '2013-12-24']
ds.sel(time=dates)
<xarray.Dataset>
Dimensions: (lat: 25, time: 3, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-07-09 2013-10-11 2013-12-24
Data variables:
air (time, lat, lon) float32 279.0 278.6 278.1 ... 296.8 296.6 296.5
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...Fancy indexing based on year, month, day, or other datetime components¶
In addition to the basic datetime indexing techniques, Xarray also supports "fancy" indexing options, which can provide more flexibility and efficiency in your data analysis tasks. You can directly access datetime components such as year, month, day, hour, etc. using the .dt accessor. Here is an example of selecting all data points from July across all years:
ds.sel(time=ds.time.dt.month == 7)
<xarray.Dataset>
Dimensions: (lat: 25, time: 248, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-07-01 ... 2014-07-31T18:00:00
Data variables:
air (time, lat, lon) float32 273.7 273.0 272.5 ... 297.5 297.6 297.8
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...Or, if you wanted to select data from a specific day of each month, you could use:
ds.sel(time=ds.time.dt.day == 15)
<xarray.Dataset>
Dimensions: (lat: 25, time: 96, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-15 ... 2014-12-15T18:00:00
Data variables:
air (time, lat, lon) float32 243.8 243.4 242.8 ... 297.1 296.9 296.9
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...Xarray Computation¶
Similar to Pandas, xarray supports different kinds of arithmetic operations
ds['air_C'] = ds['air'] - 273.15
ds['air_C']
<xarray.DataArray 'air_C' (time: 2920, lat: 25, lon: 53)>
array([[[-31.949997, -30.649994, -29.649994, ..., -40.350006,
-37.649994, -34.550003],
[-29.350006, -28.649994, -28.449997, ..., -40.350006,
-37.850006, -33.850006],
[-23.149994, -23.350006, -24.259995, ..., -39.949997,
-36.759995, -31.449997],
...,
[ 23.450012, 23.049988, 23.25 , ..., 22.25 ,
21.950012, 21.549988],
[ 22.75 , 23.049988, 23.640015, ..., 22.75 ,
22.75 , 22.049988],
[ 23.140015, 23.640015, 23.950012, ..., 23.75 ,
23.640015, 23.450012]],
[[-31.050003, -30.449997, -30.050003, ..., -41.149994,
-39.550003, -37.350006],
[-29.550003, -29.050003, -28.949997, ..., -42.149994,
-40.649994, -37.449997],
[-19.949997, -20.259995, -21.050003, ..., -42.350006,
-39.759995, -34.649994],
...
[ 20.540009, 20.73999 , 22.23999 , ..., 21.940002,
21.540009, 21.140015],
[ 23.140015, 24.040009, 24.440002, ..., 22.140015,
21.940002, 21.23999 ],
[ 24.640015, 25.23999 , 25.339996, ..., 22.540009,
22.339996, 22.040009]],
[[-28.059998, -28.86 , -29.86 , ..., -31.460007,
-31.660004, -31.36 ],
[-23.259995, -23.86 , -24.759995, ..., -33.559998,
-32.86 , -31.460007],
[-10.160004, -10.959991, -11.76001 , ..., -33.259995,
-30.559998, -26.86 ],
...,
[ 20.640015, 20.540009, 21.940002, ..., 22.140015,
21.940002, 21.540009],
[ 22.940002, 23.73999 , 24.040009, ..., 22.540009,
22.540009, 22.040009],
[ 24.540009, 24.940002, 24.940002, ..., 23.339996,
23.040009, 22.540009]]], dtype=float32)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00# Generate a boolean mask where temperature is higher than 0˚C
ds['air_C'] > 0
<xarray.DataArray 'air_C' (time: 2920, lat: 25, lon: 53)>
array([[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
...
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]]])
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00# Generate a boolean mask where long-term mean temperature is higher than 0˚C
ds['air_C'].mean(dim='time') > 0
<xarray.DataArray 'air_C' (lat: 25, lon: 53)>
array([[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]])
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0(ds['air_C'].mean(dim='time') > 0).plot()
<matplotlib.collections.QuadMesh at 0x194bbaa30>
# Apply the mask:
ds.where(ds['air_C'].mean(dim='time') > 0).mean(dim = 'time').air.plot()
<matplotlib.collections.QuadMesh at 0x194c9b100>
Broadcasting: expanding data¶
ds['air_C'].mean(dim='time').plot()
<matplotlib.collections.QuadMesh at 0x194dd1130>
(ds['air_C'].mean(dim='time')*np.cos(np.deg2rad(ds.lat))).plot()
<matplotlib.collections.QuadMesh at 0x194e8ccd0>
# Make cos(lat) two dimensional
(xr.ones_like(ds['air_C'].mean(dim='time'))*np.cos(np.deg2rad(ds.lat))).plot()
<matplotlib.collections.QuadMesh at 0x194f7a3d0>
# Make cos(lat) two dimensional
(np.cos(np.deg2rad(ds.lat))*xr.ones_like(ds.lon)).plot()
<matplotlib.collections.QuadMesh at 0x194aa2f10>
Xarray Groupby and Resample¶
groupby¶
# here's ds
ds
<xarray.Dataset>
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 241.2 242.5 243.5 ... 296.5 296.2 295.7
air_C (time, lat, lon) float32 -31.95 -30.65 -29.65 ... 23.34 23.04 22.54
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...# seasonal groups
ds.groupby("time.season")
DatasetGroupBy, grouped over 'season' 4 groups with labels 'DJF', 'JJA', 'MAM', 'SON'.
# make a seasonal mean
seasonal_mean = ds.groupby("time.season").mean()
seasonal_mean
<xarray.Dataset>
Dimensions: (lat: 25, season: 4, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* season (season) object 'DJF' 'JJA' 'MAM' 'SON'
Data variables:
air (season, lat, lon) float32 247.0 247.0 246.7 ... 299.4 299.4 299.5
air_C (season, lat, lon) float32 -26.14 -26.19 -26.43 ... 26.22 26.32
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...The seasons are out of order (they are alphabetically sorted). This is a common
annoyance. The solution is to use .sel to change the order of labels
seasonal_mean = seasonal_mean.sel(season=["DJF", "MAM", "JJA", "SON"])
seasonal_mean
<xarray.Dataset>
Dimensions: (lat: 25, season: 4, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* season (season) object 'DJF' 'MAM' 'JJA' 'SON'
Data variables:
air (season, lat, lon) float32 247.0 247.0 246.7 ... 299.4 299.4 299.5
air_C (season, lat, lon) float32 -26.14 -26.19 -26.43 ... 26.22 26.32
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...seasonal_mean.air.plot(col="season")
<xarray.plot.facetgrid.FacetGrid at 0x1950926a0>
# Make the figure to 2x2 plots:
# facet the seasonal_mean
seasonal_mean.air.plot(col="season", col_wrap=2);
# Calculate zonal average
seasonal_mean.air.mean("lon").plot.line(hue="season", y="lat");
resample¶
# resample to monthly frequency
ds.resample(time="M").mean()
<xarray.Dataset>
Dimensions: (lat: 25, time: 24, lon: 53)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-31 2013-02-28 ... 2014-12-31
Data variables:
air (time, lat, lon) float32 244.5 244.7 244.7 ... 297.7 297.7 297.7
air_C (time, lat, lon) float32 -28.68 -28.49 -28.48 ... 24.55 24.57 24.56
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...ds.resample(time="M").mean().mean(dim =['lat','lon']).air.plot()
[<matplotlib.lines.Line2D at 0x1954c0730>]